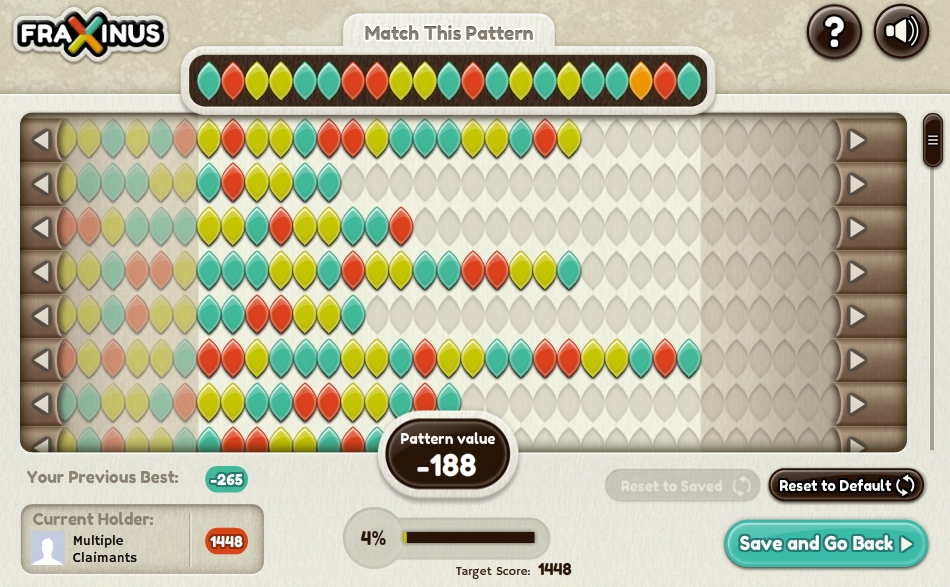
Fraxinus, the biologically, scientifically, and ecologically relevant version of Candy Crush Saga (Image Credit: Fraxinus)
In the November print issue of Discover Magazine, the article “Cooking trees to save citrus” discusses the pathogenic attack of citrus trees by a bacterium spread by Asian psyllids and how heat generation can sterilize trees from these infectious agents. Unfortunately, the ash tree, which is also under attack by a microbial pathogen, doesn’t have the same line of defense. In a race against the spread of the disease across Europe, scientists are trying to uncover the genetic mechanism for fungal resistance by recruiting the help of citizen scientists.
With autumn comes a multitude of colors—but the ash tree, faithful to its name, stays a non-conflagrant color while waiting for winter to take its leaves. Or while waiting for its microscopic predator, the Chalara fungus. Chalara causes ash dieback, a disease that has wiped out over 60% of the ash trees in Denmark, and is now sweeping across Europe, with highest incidences in Sweden and the UK. Yet some trees exhibit resilience to infection by Chalara, and it’s this curious display of biological fitness that has incited the interest of geneticists. The Sainsbury Lab (TSL), in collaboration with the John Innes Centre set about exploring this scientific problem by collecting samples from the Chalara fungus and ash trees from all over the UK, infected and resistant alike. But when confronted with the massive amount of genetic information beholden by these samples, they opted to recruit the help of citizen scientists.
When Chalara first presented itself in the UK in 2012, Dan MacLean, a professor of bioinformatics at TSL, quickly recognized that uncovering the genetic underpinnings for Chalara resistance would be challenging for current bioinformatics solutions given the complex genome structure of this fungal organism. Computational algorithms can readily sort through DNA sequence data to determine differences between sequences, but only if those sequences are of standard issue. Accordingly, MacLean and colleagues developed Fraxinus, a game named after the scientific name for the ash tree, Fraxinus excelsior, for users to play on real DNA sequence data, leveraging their unique strength over computers—visual pattern matching. Users can log in to Facebook and play against other users to match bases between multiple sequences of Chalara or ash DNA (represented as colorful leaves) in a process called sequence alignment. The player with the best alignment gets to “keep” the sequence, and the player with the most sequences wins. Easier said than done—differing bases can be challenging to resolve optimally, and meandering sequences require intricate problem solving to outcompete other players. However, Fraxinus users have played against high performance software algorithms and matched or beaten their computer opponent in more than 80% of cases.
The game just celebrated its first anniversary this August, and in the short time that the game has been hosted, over 26,000 citizen scientists have played Fraxinus, contributing to close to 3 years of continuous gameplay to better understand how ash dieback can be treated. Specifically, the output of the game helps identify regions of DNA with significant dissimilarity from one another. Alongside work to correlate these genetic differences with observations regarding ash health, Fraxinus gives scientists a narrower lens to pinpoint genes that are involved in ash dieback susceptibility.

Not So Healthy – Leaves of the Ash tree infected by the Chalara fungus
“With more accurate lists of genetic variants we will be much better placed to assess genetic variability between strains and to carry out experiments to associate particular organismal traits (such as the ability to cause or withstand infection) with their genetic root,” MacLean writes in a peer-reviewed publication in Elife. “Ultimately this will help us to breed trees that are resistant to ash dieback (or other diseases), to identify treatments, and to plan for better management of our woodlands and tree stocks.”
The Chalara genome is 60 million base pairs, the ash genome 954 million base pairs, and, putting things in perspective, the human genome is 3.3 billion base pairs in length. Compound this with a large sample size and the tens to hundreds of times each base pair has to be read to ensure high quality data, the computational problem suddenly becomes enormous. Bioinformatics solutions will one day handle all the nuanced cases that these lengthy and intricately complex genomes hold, but in the meantime, citizen scientists can progress scientists’ status of sequence analysis and have fun doing it. With Fraxinus, anyone can play for as little or as long as they like, and every game helps researchers. Interestingly, the Fraxinus platform lends itself to analyzing sequence data of any kind. So the work accomplished by TSL and their collaborating development group, Team Cooper, to develop Fraxinus could be extended to new projects for engaging citizen scientists in current genomic research problems.
“Fraxinus’ success can be solely attributed to the strength it draws from the player community,” says Shyam Rallapalli, a post-doctoral fellow who works with MacLean. “Their commitment and intelligence is vital to this research and is an extremely valuable part of what we do.”
It’s unclear how infectivity of Chalara came to the European continent and whether anthropogenic activity has contributed to its spread. However, human intervention to better understand the mechanism by which Chalara infects its host and how the ash tree can fight back will be important for ecological stability as well as industrial stability. The UK alone stands to lose 90-95% of its ash trees in the coming years if Chalara remains unchecked.
So this fall, get online and use your visual discerning skills on Fraxinus to swipe right, and swipe left (albeit moving patterns to align sequence data, not people’s faces) and help scientists uncover clues in the race to to save the European ash tree.
References
MacClean. Cutting edge: Changing the rules of the game. Elife 2:e01294 (2013).
***
Fraxinus is one of more than 800 projects on SciStarter, an online citizen science hotspot. Use our project finder to search adn participate in citizen science that interests you!